
Compounding (grouping) of LC-MS features
Source:vignettes/LC-MS-feature-grouping.Rmd
LC-MS-feature-grouping.RmdPackage: xcms
Authors: Johannes Rainer
Modified: 2026-02-19 12:46:13.091735
Compiled: Thu Feb 19 13:35:58 2026
Introduction
In a typical LC-MS-based metabolomics experiment compounds eluting from the chromatography are first ionized before being measured by mass spectrometry (MS). During the ionization different (multiple) ions can be generated from the same compound which all will be measured by MS. In general, the resulting data is then pre-processed to identify chromatographic peaks in the data and to group these across samples in the correspondence analysis. The result are distinct LC-MS features, characterized by their specific m/z and retention time range. Different ions generated during ionization will be detected as different features. Compounding aims now at grouping such features presumably representing signal from the same originating compound to reduce data set complexity (and to aid in subsequent annotation steps). General MS feature grouping functionality if defined by the MsFeatures package with additional functionality being implemented in the xcms package to enable the compounding of LC-MS data.
This document provides a simple compounding workflow using xcms. Note that the present functionality does not (yet) aggregate or combine the actual features per values, but does only define the feature groups (one per compound).
Compounding of LC-MS data
We demonstrate the compounding (feature grouping) functionality on the simple toy data set used also in the xcms package and provided through the faahKO package. This data set consists of samples from 4 mice with knock-out of the fatty acid amide hydrolase (FAAH) and 4 wild type mice. Pre-processing of this data set is described in detail in the main vignette of the xcms package. Below we load all required packages and the result from this pre-processing updating also the location of the respective raw data files on the current machine.
library(MSnbase)
library(xcms)
library(faahKO)
library(MsFeatures)
xmse <- loadXcmsData("xmse")Before performing the feature grouping we inspect the result object.
With featureDefinitions we can extract the results from the
correspondence analysis.
featureDefinitions(xmse) |> head()## mzmed mzmin mzmax rtmed rtmin rtmax npeaks KO WT peakidx
## FT001 200.1 200.1 200.1 2902.634 2882.603 2922.664 2 2 0 458, 116....
## FT002 205.0 205.0 205.0 2789.901 2782.955 2796.531 8 4 4 44, 443,....
## FT003 206.0 206.0 206.0 2789.405 2781.389 2794.219 7 3 4 29, 430,....
## FT004 207.1 207.1 207.1 2718.560 2714.047 2727.347 7 4 3 16, 420,....
## FT005 233.0 233.0 233.1 3023.579 3015.145 3043.959 7 3 4 69, 959,....
## FT006 241.1 241.1 241.2 3683.299 3661.586 3695.886 8 3 4 276, 284....
## ms_level
## FT001 1
## FT002 1
## FT003 1
## FT004 1
## FT005 1
## FT006 1Each row in this data frame represents the definition of one feature,
with its average and range of m/z and retention time. Column
"peakidx" provides the index of each chromatographic peak
which is assigned to the feature in the chromPeaks matrix
of the result object. The featureValues function allows to
extract feature values, i.e. a matrix with feature abundances,
one row per feature and columns representing the samples of the present
data set.
Below we extract the feature values with and without filled-in peak data. Without the gap-filled data only abundances from detected chromatographic peaks are reported. In the gap-filled data, for samples in which no chromatographic peak for a feature was detected, all signal from the m/z - retention time range defined based on the detected chromatographic peaks was integrated.
head(featureValues(xmse, filled = FALSE))## ko15.CDF ko16.CDF ko21.CDF ko22.CDF wt15.CDF wt16.CDF wt21.CDF
## FT001 NA 506848.9 NA 169955.6 NA NA NA
## FT002 1924712.0 1757151.0 1383416.7 1180288.2 2129885.1 1634342.0 1623589.2
## FT003 213659.3 289500.7 NA 178285.7 253825.6 241844.4 240606.0
## FT004 349011.5 451863.7 343897.8 208002.8 364609.8 360908.9 NA
## FT005 286221.4 NA 164009.0 149097.6 255697.7 311296.8 366441.5
## FT006 1160580.5 NA 380970.3 588986.4 1286883.0 1739516.6 639755.3
## wt22.CDF
## FT001 NA
## FT002 1354004.9
## FT003 185399.5
## FT004 221937.5
## FT005 271128.0
## FT006 508546.4
head(featureValues(xmse, filled = TRUE))## ko15.CDF ko16.CDF ko21.CDF ko22.CDF wt15.CDF wt16.CDF wt21.CDF
## FT001 135162.4 506848.9 111657.3 169955.6 209929.4 141607.9 226853.7
## FT002 1924712.0 1757151.0 1383416.7 1180288.2 2129885.1 1634342.0 1623589.2
## FT003 213659.3 289500.7 164380.7 178285.7 253825.6 241844.4 240606.0
## FT004 349011.5 451863.7 343897.8 208002.8 364609.8 360908.9 226234.4
## FT005 286221.4 285857.6 164009.0 149097.6 255697.7 311296.8 366441.5
## FT006 1160580.5 1102832.6 380970.3 588986.4 1286883.0 1739516.6 639755.3
## wt22.CDF
## FT001 138341.2
## FT002 1354004.9
## FT003 185399.5
## FT004 221937.5
## FT005 271128.0
## FT006 508546.4In total 351 features have been defined in the present data set, many of which most likely represent signal from different ions (adducts or isotopes) of the same compound. The aim of the grouping functions of are now to define which features most likely come from the same original compound. The feature grouping functions base on the following assumptions/properties of LC-MS data:
- Features (ions) of the same compound should have similar retention time.
- The abundance of features (ions) of the same compound should have a similar pattern across samples, i.e. if a compound is highly concentrated in one sample and low in another, all ions from it should follow the same pattern.
- The peak shape of extracted ion chromatograms (EIC) of features of the same compound should be similar as it should follow the elution pattern of the original compound from the LC.
The main method to perform the feature grouping is called
groupFeatures which takes an xcms result object
(i.e., an XcmsExperiment or XCMSnExp) as input
as well as a parameter object to chose the grouping algorithm and
specify its settings. xcms provides and supports the following
grouping approaches:
-
SimilarRtimeParam: perform an initial grouping based on similar retention time. -
AbundanceSimilarityParam: perform a feature grouping based on correlation of feature abundances (values) across samples. -
EicSimilarityParam: perform a feature grouping based on correlation of EICs.
Calling groupFeatures on an xcms result object
will perform a feature grouping assigning each feature in the data set
to a feature group. These feature groups are stored as an
additional column called "feature_group" in the
featureDefinition data frame of the result object and can
be accessed with the featureGroups function. Any subsequent
groupFeature call will sub-group (refine) the
identified feature groups further. It is thus possible to use a single
grouping approach, or to combine multiple of them to generate the
desired feature grouping in an incremental fashion. While the individual
feature grouping algorithms can be called in any order, it is advisable
to use the EicSimilarityParam as last refinement step,
because it is computationally very expensive, especially if applied to a
result object without pre-defined feature groups or if the feature
groups are very large.
Grouping of features by similar retention time
The most intuitive and simple way to group features is based on their retention time. Before we perform this initial grouping we evaluate retention times and m/z of all features in the present data set.
plot(featureDefinitions(xmse)$rtmed, featureDefinitions(xmse)$mzmed,
xlab = "retention time", ylab = "m/z", main = "features",
col = "#00000080", pch = 21, bg = "#00000040")
grid()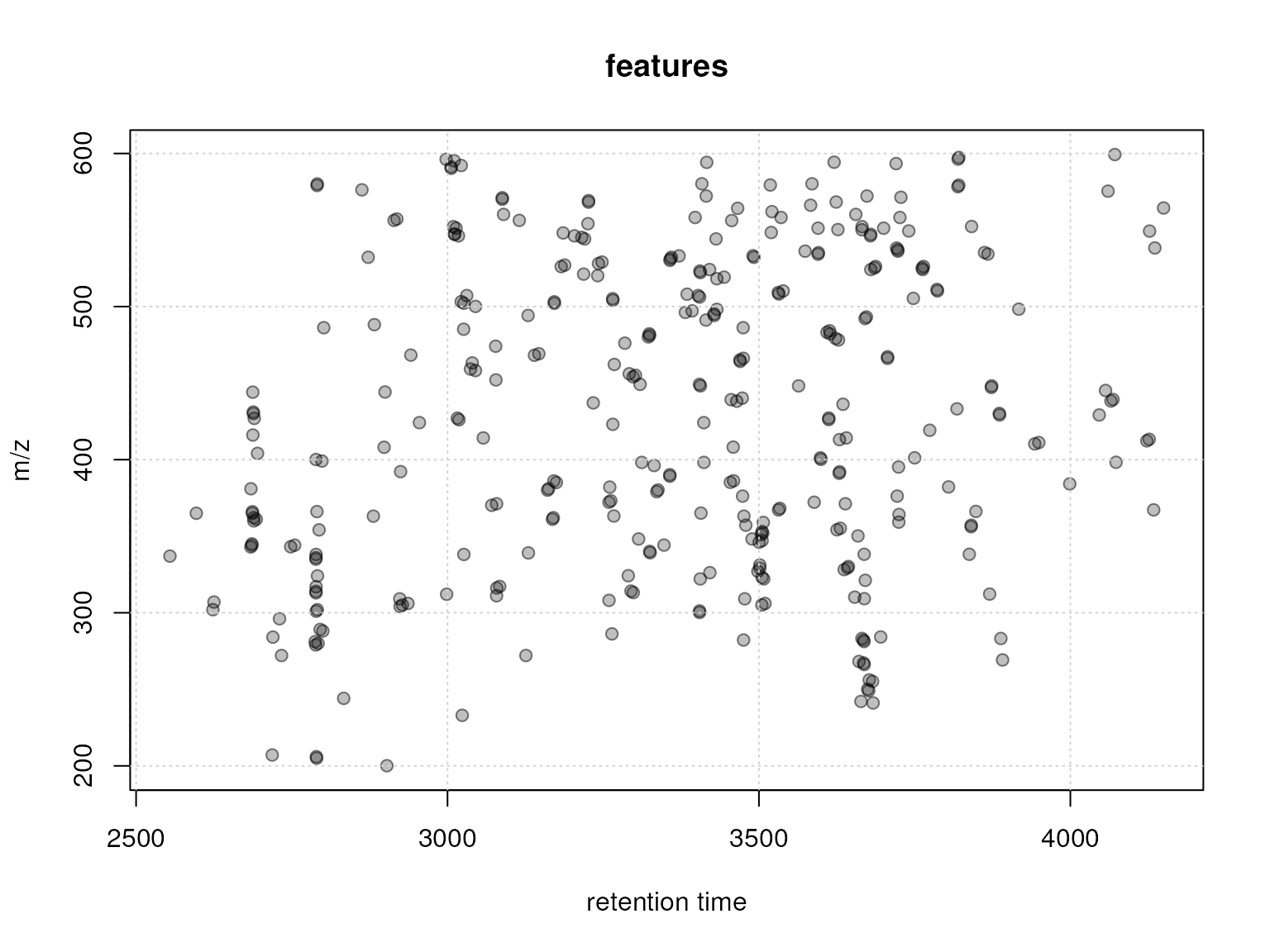
Plot of retention times and m/z for all features in the data set.
Several features with about the same retention time (but different m/z) can be spotted, especially at the beginning of the LC. We thus below group features within a retention time window of 10 seconds into feature groups.
xmse <- groupFeatures(xmse, param = SimilarRtimeParam(10))The results from the feature grouping can be accessed with the
featureGroups function. Below we determine the size of each
of these feature groups (i.e. how many features are grouped
together).
table(featureGroups(xmse))##
## FG.001 FG.002 FG.003 FG.004 FG.005 FG.006 FG.007 FG.008 FG.009 FG.010 FG.011
## 4 6 2 4 6 5 2 4 3 4 3
## FG.012 FG.013 FG.014 FG.015 FG.016 FG.017 FG.018 FG.019 FG.020 FG.021 FG.022
## 6 4 2 3 3 3 4 3 2 5 2
## FG.023 FG.024 FG.025 FG.026 FG.027 FG.028 FG.029 FG.030 FG.031 FG.032 FG.033
## 2 4 5 2 5 2 3 4 2 2 4
## FG.034 FG.035 FG.036 FG.037 FG.038 FG.039 FG.040 FG.041 FG.042 FG.043 FG.044
## 2 2 6 3 2 2 4 2 2 4 4
## FG.045 FG.046 FG.047 FG.048 FG.049 FG.050 FG.051 FG.052 FG.053 FG.054 FG.055
## 2 3 2 3 3 5 2 3 4 2 6
## FG.056 FG.057 FG.058 FG.059 FG.060 FG.061 FG.062 FG.063 FG.064 FG.065 FG.066
## 2 6 4 2 4 3 3 2 2 2 2
## FG.067 FG.068 FG.069 FG.070 FG.071 FG.072 FG.073 FG.074 FG.075 FG.076 FG.077
## 3 2 2 3 2 3 2 3 3 3 3
## FG.078 FG.079 FG.080 FG.081 FG.082 FG.083 FG.084 FG.085 FG.086 FG.087 FG.088
## 2 3 3 3 2 3 3 3 3 3 2
## FG.089 FG.090 FG.091 FG.092 FG.093 FG.094 FG.095 FG.096 FG.097 FG.098 FG.099
## 3 2 2 2 2 3 2 4 2 3 3
## FG.100 FG.101 FG.102 FG.103 FG.104 FG.105 FG.106 FG.107 FG.108 FG.109 FG.110
## 3 3 2 2 2 2 2 2 2 2 2
## FG.111 FG.112 FG.113 FG.114 FG.115 FG.116 FG.117 FG.118 FG.119 FG.120 FG.121
## 2 2 1 1 1 1 1 1 1 1 1
## FG.122 FG.123 FG.124 FG.125 FG.126 FG.127 FG.128 FG.129 FG.130 FG.131 FG.132
## 1 1 1 1 1 1 1 1 1 1 1
## FG.133
## 1In addition we visualize these feature groups with the
plotFeatureGroups function which shows all features in the
m/z - retention time space with grouped features being connected with a
line.
plotFeatureGroups(xmse, pch = 21, lwd = 2, col = "#00000040",
bg = "#00000020")
grid()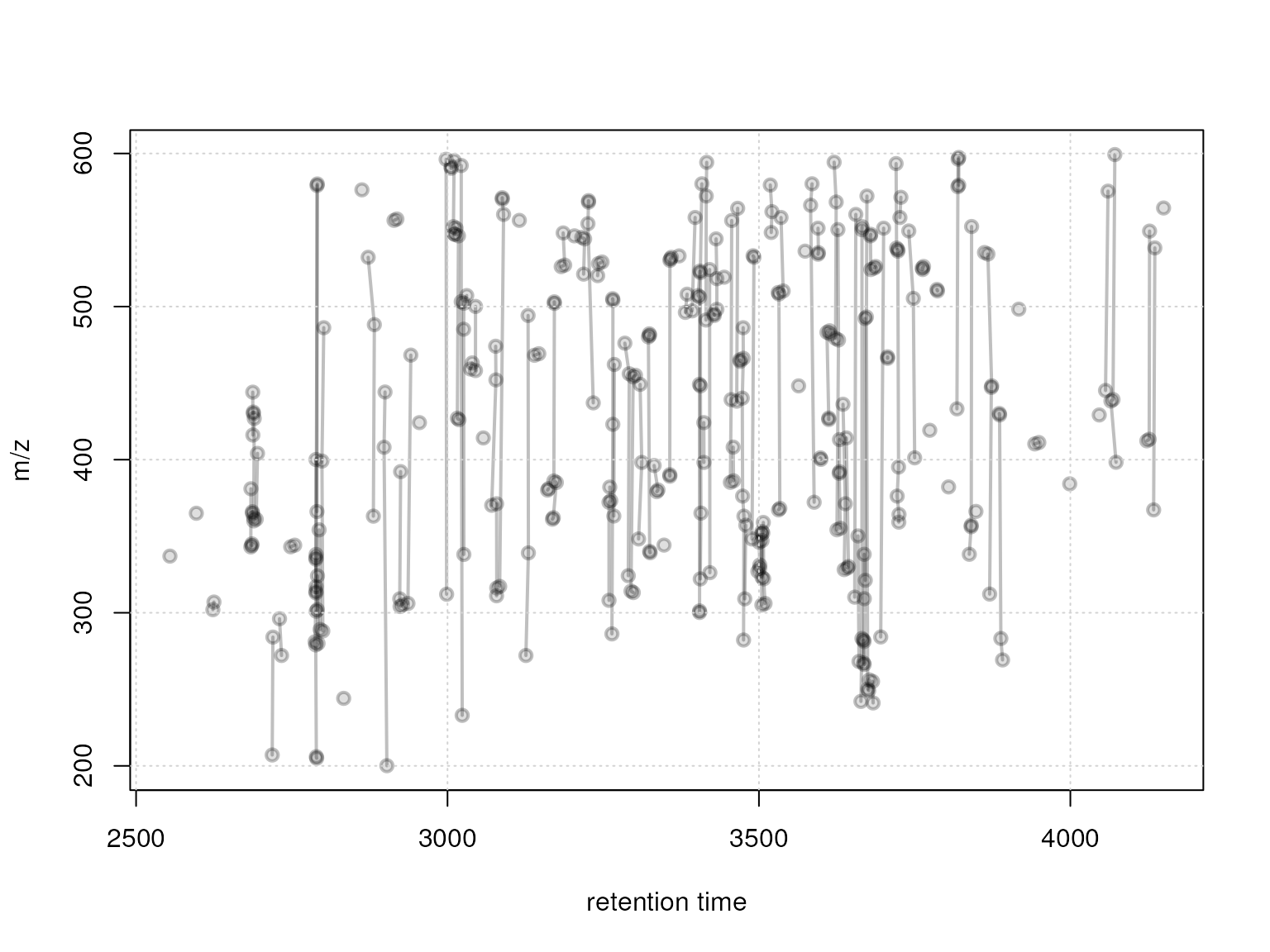
Feature groups defined with a rt window of 10 seconds
Let’s assume we don’t agree with this feature grouping, also knowing
that there were quite large shifts in retention times between runs. We
thus first remove any defined feature groups assigning them a value of
NULL and then re-perform the feature grouping using a
larger rt window.
## Remove previous feature grouping results to repeat the rtime-based
## feature grouping with different setting
featureDefinitions(xmse)$feature_group <- NULL
## Repeat the grouping
xmse <- groupFeatures(xmse, SimilarRtimeParam(20))
table(featureGroups(xmse))##
## FG.001 FG.002 FG.003 FG.004 FG.005 FG.006 FG.007 FG.008 FG.009 FG.010 FG.011
## 4 6 2 4 6 5 2 4 3 4 3
## FG.012 FG.013 FG.014 FG.015 FG.016 FG.017 FG.018 FG.019 FG.020 FG.021 FG.022
## 6 4 2 3 3 3 4 3 2 5 2
## FG.023 FG.024 FG.025 FG.026 FG.027 FG.028 FG.029 FG.030 FG.031 FG.032 FG.033
## 2 4 6 3 5 2 3 4 2 2 4
## FG.034 FG.035 FG.036 FG.037 FG.038 FG.039 FG.040 FG.041 FG.042 FG.043 FG.044
## 2 2 6 3 2 2 4 2 2 4 5
## FG.045 FG.046 FG.047 FG.048 FG.049 FG.050 FG.051 FG.052 FG.053 FG.054 FG.055
## 2 3 2 3 4 5 2 4 4 2 6
## FG.056 FG.057 FG.058 FG.059 FG.060 FG.061 FG.062 FG.063 FG.064 FG.065 FG.066
## 2 6 4 2 5 3 3 2 2 2 2
## FG.067 FG.068 FG.069 FG.070 FG.071 FG.072 FG.073 FG.074 FG.075 FG.076 FG.077
## 4 2 2 3 2 3 2 4 3 3 3
## FG.078 FG.079 FG.080 FG.081 FG.082 FG.083 FG.084 FG.085 FG.086 FG.087 FG.088
## 2 3 3 3 2 3 3 4 3 4 2
## FG.089 FG.090 FG.091 FG.092 FG.093 FG.094 FG.095 FG.096 FG.097 FG.098 FG.099
## 3 2 2 2 2 3 3 4 2 3 4
## FG.100 FG.101 FG.102 FG.103 FG.104 FG.105 FG.106 FG.107 FG.108 FG.109 FG.110
## 3 3 3 2 2 3 2 3 2 2 2
## FG.111 FG.112 FG.113 FG.114 FG.115 FG.116 FG.117 FG.118
## 2 2 1 1 1 1 1 1
plotFeatureGroups(xmse, pch = 21, lwd = 2, col = "#00000040", bg = "#00000020")
grid()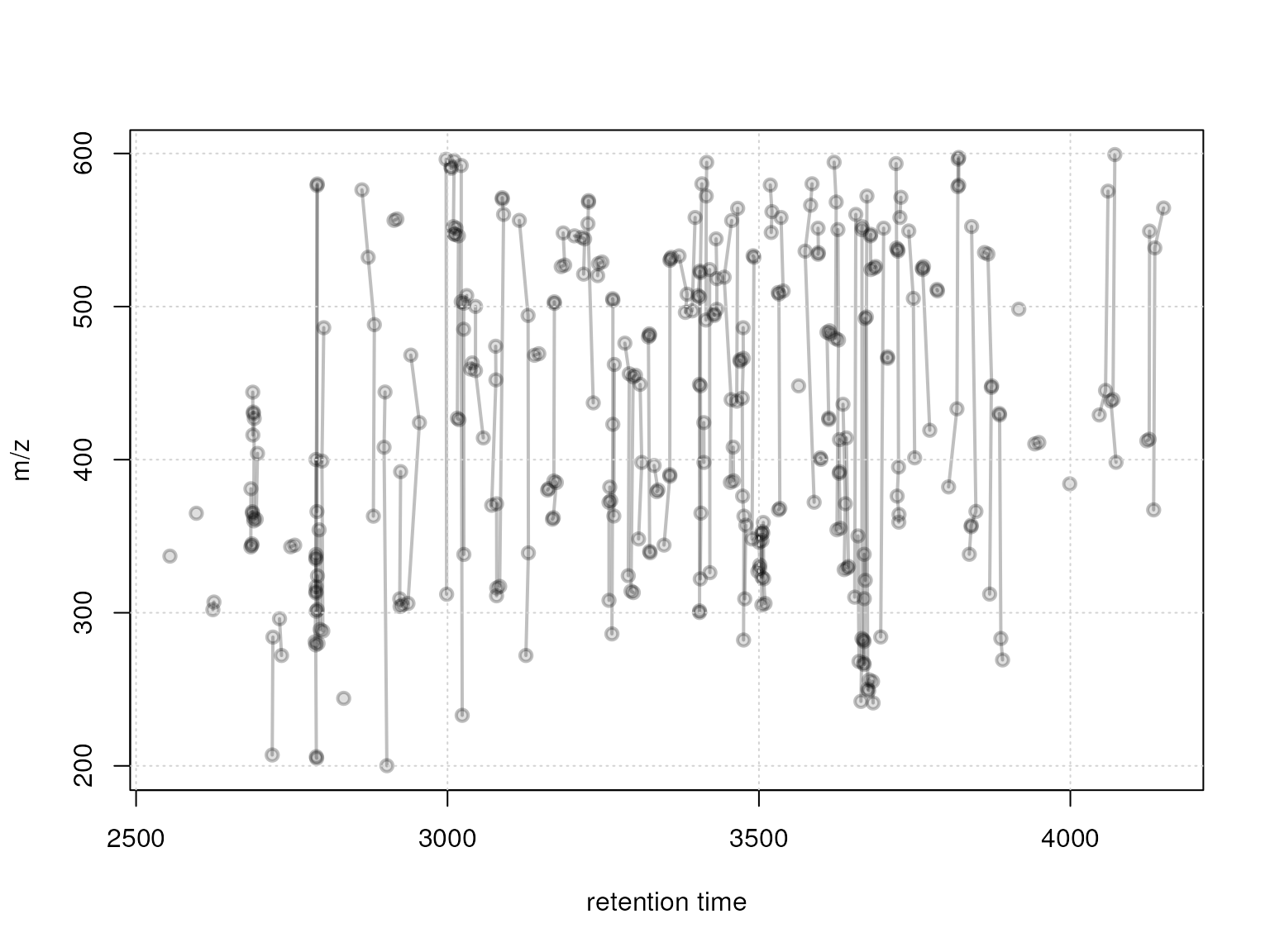
Feature groups defined with a rt window of 20 seconds
Grouping by similar retention time grouped the in total 351 features into 118 feature groups.
Grouping of features by abundance correlation across samples
Assuming we are OK with the crude initial feature grouping
from the previous section, we can next refine the feature
groups considering also the feature abundances across samples. We can
use the groupFeatures method with an
AbundanceSimilarityParam object. This approach performs a
pairwise correlation between all (non-missing) feature values
(abundances; across samples) between all features of a predefined
feature group (such as defined in the previous section). Features that
have a correlation >= threshold are grouped together.
Feature grouping based on this approach works best for features with a
higher variability in their concentration across samples. Parameter
subset allows to restrict the analysis to a subset of
samples and allows thus to e.g. exclude QC sample pools from this
correlation as these could artificially increase the correlation. Other
parameters are passed directly to the internal
featureValues call that extracts the feature values on
which the correlation should be performed.
Before performing the grouping we could also evaluate the correlation of features based on their (log2 transformed) abundances across samples with a heatmap.
library(pheatmap)
fvals <- log2(featureValues(xmse, filled = TRUE))
cormat <- cor(t(fvals), use = "pairwise.complete.obs")
ann <- data.frame(fgroup = featureGroups(xmse))
rownames(ann) <- rownames(cormat)
res <- pheatmap(cormat, annotation_row = ann, cluster_rows = TRUE,
cluster_cols = TRUE, show_rownames = FALSE,
show_colnames = FALSE)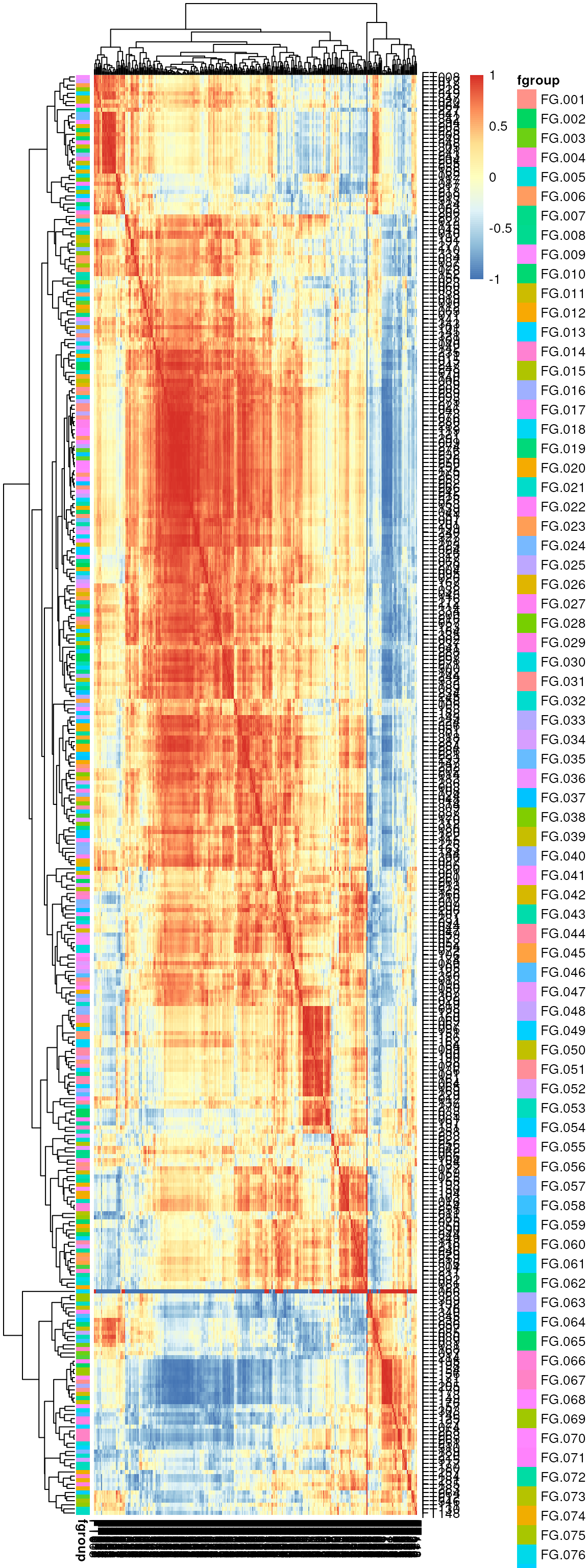
Heatmap representing pairwise correlation of features based on abundances across samples. Features are grouped on rows and columns.
Some large correlations can be observed for several groups of features, but many of them are not within the same feature group defined in the previous section (i.e. are not eluting at the same time). These might thus represent correlated metabolites, but not ions or adducts from the same metabolite.
Below we use the groupFeatures with the
AbundanceSimilarityParam to group features with a
correlation coefficient higher than 0.7 including both detected and
filled-in signal. Whether filled-in or only detected signal should be
used in the correlation analysis should be evaluated from data set to
data set. By specifying transform = log2 we tell the
function to log2 transform the abundance prior to the correlation
analysis. See the help page for groupFeatures with
AbundanceSimilarityParam in the xcms package
for details and options.
xmse <- groupFeatures(
xmse, AbundanceSimilarityParam(threshold = 0.7, transform = log2),
filled = TRUE)
table(featureGroups(xmse))##
## FG.001.001 FG.001.002 FG.002.001 FG.002.002 FG.002.003 FG.002.004 FG.003.001
## 3 1 2 2 1 1 1
## FG.003.002 FG.004.001 FG.004.002 FG.004.003 FG.005.001 FG.005.002 FG.005.003
## 1 2 1 1 3 2 1
## FG.006.001 FG.006.002 FG.007.001 FG.008.001 FG.008.002 FG.008.003 FG.008.004
## 4 1 2 1 1 1 1
## FG.009.001 FG.010.001 FG.010.002 FG.011.001 FG.011.002 FG.012.001 FG.012.002
## 3 3 1 2 1 3 2
## FG.012.003 FG.013.001 FG.013.002 FG.014.001 FG.015.001 FG.015.002 FG.015.003
## 1 3 1 2 1 1 1
## FG.016.001 FG.016.002 FG.016.003 FG.017.001 FG.017.002 FG.018.001 FG.018.002
## 1 1 1 2 1 2 2
## FG.019.001 FG.019.002 FG.019.003 FG.020.001 FG.020.002 FG.021.001 FG.021.002
## 1 1 1 1 1 1 1
## FG.021.003 FG.021.004 FG.021.005 FG.022.001 FG.022.002 FG.023.001 FG.024.001
## 1 1 1 1 1 2 2
## FG.024.002 FG.025.001 FG.025.002 FG.025.003 FG.026.001 FG.026.002 FG.027.001
## 2 4 1 1 2 1 2
## FG.027.002 FG.027.003 FG.027.004 FG.028.001 FG.028.002 FG.029.001 FG.029.002
## 1 1 1 1 1 1 1
## FG.029.003 FG.030.001 FG.030.002 FG.031.001 FG.032.001 FG.032.002 FG.033.001
## 1 3 1 2 1 1 2
## FG.033.002 FG.033.003 FG.034.001 FG.034.002 FG.035.001 FG.036.001 FG.036.002
## 1 1 1 1 2 3 1
## FG.036.003 FG.036.004 FG.037.001 FG.038.001 FG.038.002 FG.039.001 FG.039.002
## 1 1 3 1 1 1 1
## FG.040.001 FG.040.002 FG.040.003 FG.040.004 FG.041.001 FG.041.002 FG.042.001
## 1 1 1 1 1 1 1
## FG.042.002 FG.043.001 FG.043.002 FG.044.001 FG.044.002 FG.045.001 FG.046.001
## 1 3 1 3 2 2 1
## FG.046.002 FG.046.003 FG.047.001 FG.047.002 FG.048.001 FG.048.002 FG.049.001
## 1 1 1 1 2 1 1
## FG.049.002 FG.049.003 FG.049.004 FG.050.001 FG.050.002 FG.051.001 FG.051.002
## 1 1 1 4 1 1 1
## FG.052.001 FG.052.002 FG.052.003 FG.052.004 FG.053.001 FG.053.002 FG.054.001
## 1 1 1 1 2 2 1
## FG.054.002 FG.055.001 FG.056.001 FG.057.001 FG.057.002 FG.057.003 FG.057.004
## 1 6 2 2 1 1 1
## FG.057.005 FG.058.001 FG.058.002 FG.058.003 FG.058.004 FG.059.001 FG.060.001
## 1 1 1 1 1 2 2
## FG.060.002 FG.060.003 FG.061.001 FG.061.002 FG.062.001 FG.062.002 FG.063.001
## 2 1 2 1 2 1 1
## FG.063.002 FG.064.001 FG.065.001 FG.065.002 FG.066.001 FG.067.001 FG.067.002
## 1 2 1 1 2 3 1
## FG.068.001 FG.068.002 FG.069.001 FG.070.001 FG.070.002 FG.071.001 FG.071.002
## 1 1 2 2 1 1 1
## FG.072.001 FG.072.002 FG.072.003 FG.073.001 FG.073.002 FG.074.001 FG.074.002
## 1 1 1 1 1 2 1
## FG.074.003 FG.075.001 FG.075.002 FG.075.003 FG.076.001 FG.076.002 FG.076.003
## 1 1 1 1 1 1 1
## FG.077.001 FG.077.002 FG.078.001 FG.079.001 FG.079.002 FG.079.003 FG.080.001
## 2 1 2 1 1 1 2
## FG.080.002 FG.081.001 FG.081.002 FG.082.001 FG.082.002 FG.083.001 FG.083.002
## 1 2 1 1 1 1 1
## FG.083.003 FG.084.001 FG.084.002 FG.084.003 FG.085.001 FG.085.002 FG.086.001
## 1 1 1 1 3 1 2
## FG.086.002 FG.087.001 FG.087.002 FG.088.001 FG.088.002 FG.089.001 FG.089.002
## 1 2 2 1 1 2 1
## FG.090.001 FG.090.002 FG.091.001 FG.092.001 FG.093.001 FG.094.001 FG.094.002
## 1 1 2 2 2 2 1
## FG.095.001 FG.096.001 FG.096.002 FG.097.001 FG.097.002 FG.098.001 FG.098.002
## 3 3 1 1 1 1 1
## FG.098.003 FG.099.001 FG.099.002 FG.099.003 FG.100.001 FG.100.002 FG.101.001
## 1 2 1 1 2 1 3
## FG.102.001 FG.102.002 FG.102.003 FG.103.001 FG.104.001 FG.104.002 FG.105.001
## 1 1 1 2 1 1 2
## FG.105.002 FG.106.001 FG.107.001 FG.107.002 FG.108.001 FG.108.002 FG.109.001
## 1 2 2 1 1 1 1
## FG.109.002 FG.110.001 FG.111.001 FG.112.001 FG.112.002 FG.113.001 FG.114.001
## 1 2 2 1 1 1 1
## FG.115.001 FG.116.001 FG.117.001 FG.118.001
## 1 1 1 1Many of the larger retention time-based feature groups have been
splitted into two or more sub-groups based on the correlation of their
feature abundances (e.g. the feature group FG.004 has been
further split into feature groups FG.004.001,
FG.004.002 and FG.004.003). We evaluate this further
refinement for feature group "FG.040" by plotting their
pairwise correlation. To better visualize the feature grouping we in
addition define a custom panel plot for the pairs
function that plots data points in blue for features with a correlation
coefficient above the selected threshold (0.7) and red
otherwise.
cor_plot <- function(x, y) {
C <- cor(x, y, use = "pairwise.complete.obs")
col <- ifelse(C >= 0.7, yes = "#0000ff80", no = "#ff000080")
points(x, y, pch = 16, col = col)
grid()
}
fts <- grep("FG.040", featureGroups(xmse))
pairs(t(fvals[fts, ]), gap = 0.1, main = "FG.040", panel = cor_plot)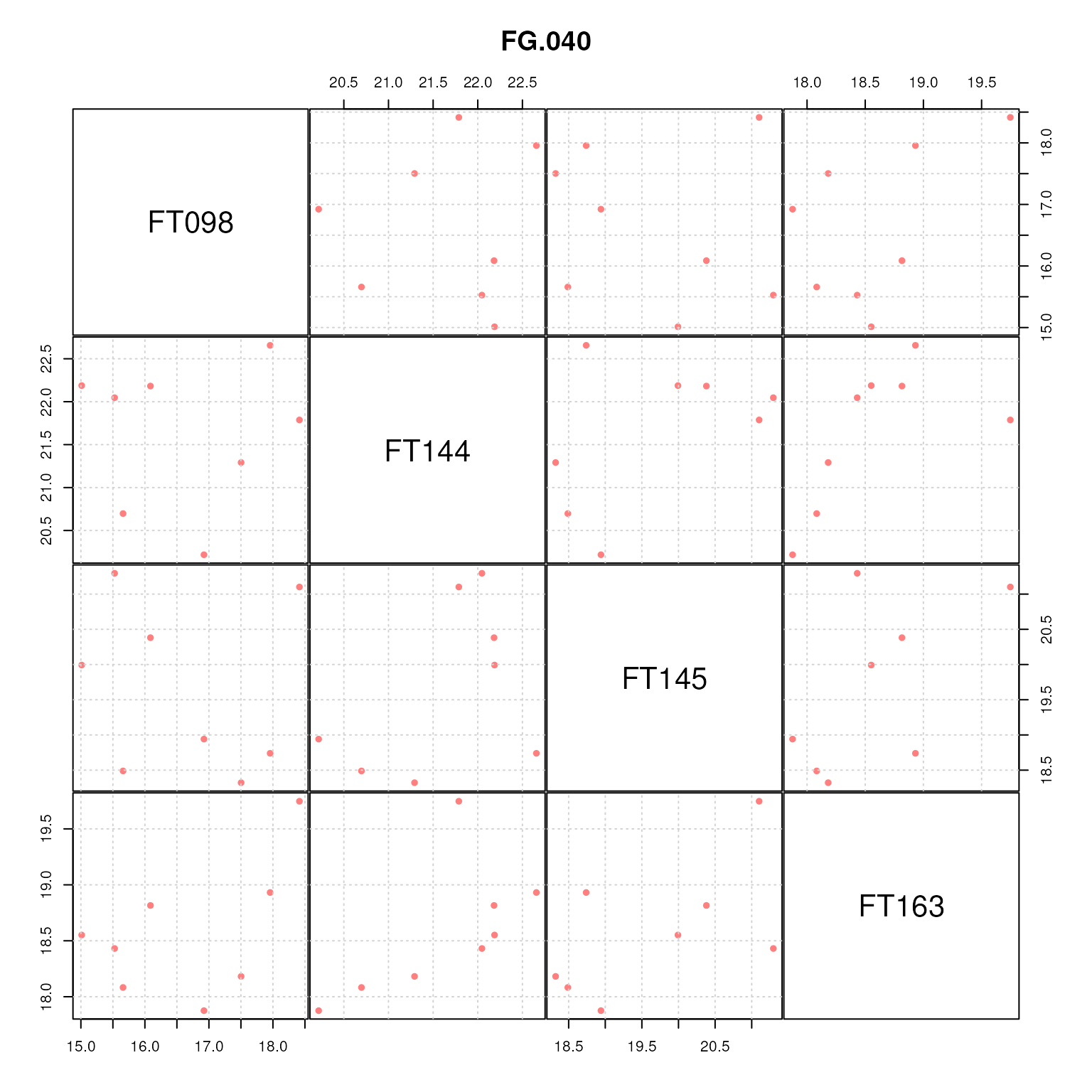
Pairwise correlation plot for all features initially grouped into the feature group FG.040.
Indeed, correlation seems only to be present between some of the features in this retention time feature group, e.g. clearly between FT273 and FT274 and also between FT143 and FT273. Note however that this abundance correlation suffers from relatively few samples (8 in total), and a relatively small variance in abundances across these samples.
After feature grouping by abundance correlation, the 351 features have been grouped into 249 feature groups.
Grouping of features by similarity of their EICs
The chromatographic peak shape of an ion of a compound should be
highly similar to the elution pattern of this compound. Thus, features
from the same compound are assumed to have similar peak shapes of their
EICs within the same sample. A grouping of features
based on similarity of their EICs can be performed with the
groupFeatures and the EicSimilarityParam
object. It is advisable to perform the peak shape correlation only on a
subset of samples (because peak shape correlation is computationally
intense and because chromatographic peaks of low intensity features are
notoriously noisy). The EicSimilarityParam approach allows
to select the number of top n samples (ordered by total
intensity of feature abundances per feature group) on which the
correlation should be performed with parameter n. With
n =3, the 3 samples with the highest signal for all
features in a respective feature group will be first identified and then
a pairwise similarity calculation will be performed on the EICs between
each of these samples. The resulting similarity score from these 3
samples will then be aggregated into a single score by taking the 75%
quantile across the 3 samples. This value is then subsequently compared
with the cut-off for similarity (parameter threshold) and
features with a score >= threshold are grouped into the
same feature group.
Below we group the features based on similarity of their EICs in the
two samples with the highest total signal for the respective feature
groups. By default, a Pearson correlation coefficient is used as
similarity score but any similarity/distance metric function could be
used instead (parameter FUN of the
EicSimilarityParam - see the respective help page
?EicSimilarityParam for details and options). We define as
a threshold a correlation coefficient of 0.7.
xmse <- groupFeatures(xmse, EicSimilarityParam(threshold = 0.7, n = 2))This is the computationally most intense approach since it involves also loading the raw MS data to extract the ion chromatograms for each feature. The results of the grouping are shown below.
table(featureGroups(xmse))##
## FG.001.001.001 FG.001.002.001 FG.002.001.001 FG.002.002.001 FG.002.003.001
## 3 1 2 2 1
## FG.002.004.001 FG.003.001.001 FG.003.002.001 FG.004.001.001 FG.004.002.001
## 1 1 1 2 1
## FG.004.003.001 FG.005.001.001 FG.005.002.001 FG.005.003.001 FG.006.001.001
## 1 3 2 1 4
## FG.006.002.001 FG.007.001.001 FG.007.001.002 FG.008.001.001 FG.008.002.001
## 1 1 1 1 1
## FG.008.003.001 FG.008.004.001 FG.009.001.001 FG.010.001.001 FG.010.002.001
## 1 1 3 3 1
## FG.011.001.001 FG.011.002.001 FG.012.001.001 FG.012.002.001 FG.012.003.001
## 2 1 3 2 1
## FG.013.001.001 FG.013.002.001 FG.014.001.001 FG.015.001.001 FG.015.002.001
## 3 1 2 1 1
## FG.015.003.001 FG.016.001.001 FG.016.002.001 FG.016.003.001 FG.017.001.001
## 1 1 1 1 2
## FG.017.002.001 FG.018.001.001 FG.018.002.001 FG.019.001.001 FG.019.002.001
## 1 2 2 1 1
## FG.019.003.001 FG.020.001.001 FG.020.002.001 FG.021.001.001 FG.021.002.001
## 1 1 1 1 1
## FG.021.003.001 FG.021.004.001 FG.021.005.001 FG.022.001.001 FG.022.002.001
## 1 1 1 1 1
## FG.023.001.001 FG.024.001.001 FG.024.001.002 FG.024.002.001 FG.025.001.001
## 2 1 1 2 4
## FG.025.002.001 FG.025.003.001 FG.026.001.001 FG.026.001.002 FG.026.002.001
## 1 1 1 1 1
## FG.027.001.001 FG.027.002.001 FG.027.003.001 FG.027.004.001 FG.028.001.001
## 2 1 1 1 1
## FG.028.002.001 FG.029.001.001 FG.029.002.001 FG.029.003.001 FG.030.001.001
## 1 1 1 1 2
## FG.030.001.002 FG.030.002.001 FG.031.001.001 FG.032.001.001 FG.032.002.001
## 1 1 2 1 1
## FG.033.001.001 FG.033.002.001 FG.033.003.001 FG.034.001.001 FG.034.002.001
## 2 1 1 1 1
## FG.035.001.001 FG.036.001.001 FG.036.001.002 FG.036.002.001 FG.036.003.001
## 2 2 1 1 1
## FG.036.004.001 FG.037.001.001 FG.037.001.002 FG.038.001.001 FG.038.002.001
## 1 2 1 1 1
## FG.039.001.001 FG.039.002.001 FG.040.001.001 FG.040.002.001 FG.040.003.001
## 1 1 1 1 1
## FG.040.004.001 FG.041.001.001 FG.041.002.001 FG.042.001.001 FG.042.002.001
## 1 1 1 1 1
## FG.043.001.001 FG.043.002.001 FG.044.001.001 FG.044.001.002 FG.044.002.001
## 3 1 2 1 1
## FG.044.002.002 FG.045.001.001 FG.046.001.001 FG.046.002.001 FG.046.003.001
## 1 2 1 1 1
## FG.047.001.001 FG.047.002.001 FG.048.001.001 FG.048.002.001 FG.049.001.001
## 1 1 2 1 1
## FG.049.002.001 FG.049.003.001 FG.049.004.001 FG.050.001.001 FG.050.001.002
## 1 1 1 3 1
## FG.050.002.001 FG.051.001.001 FG.051.002.001 FG.052.001.001 FG.052.002.001
## 1 1 1 1 1
## FG.052.003.001 FG.052.004.001 FG.053.001.001 FG.053.002.001 FG.054.001.001
## 1 1 2 2 1
## FG.054.002.001 FG.055.001.001 FG.056.001.001 FG.056.001.002 FG.057.001.001
## 1 6 1 1 2
## FG.057.002.001 FG.057.003.001 FG.057.004.001 FG.057.005.001 FG.058.001.001
## 1 1 1 1 1
## FG.058.002.001 FG.058.003.001 FG.058.004.001 FG.059.001.001 FG.060.001.001
## 1 1 1 2 2
## FG.060.002.001 FG.060.003.001 FG.061.001.001 FG.061.002.001 FG.062.001.001
## 2 1 2 1 2
## FG.062.002.001 FG.063.001.001 FG.063.002.001 FG.064.001.001 FG.065.001.001
## 1 1 1 2 1
## FG.065.002.001 FG.066.001.001 FG.067.001.001 FG.067.002.001 FG.068.001.001
## 1 2 3 1 1
## FG.068.002.001 FG.069.001.001 FG.070.001.001 FG.070.002.001 FG.071.001.001
## 1 2 2 1 1
## FG.071.002.001 FG.072.001.001 FG.072.002.001 FG.072.003.001 FG.073.001.001
## 1 1 1 1 1
## FG.073.002.001 FG.074.001.001 FG.074.002.001 FG.074.003.001 FG.075.001.001
## 1 2 1 1 1
## FG.075.002.001 FG.075.003.001 FG.076.001.001 FG.076.002.001 FG.076.003.001
## 1 1 1 1 1
## FG.077.001.001 FG.077.002.001 FG.078.001.001 FG.079.001.001 FG.079.002.001
## 2 1 2 1 1
## FG.079.003.001 FG.080.001.001 FG.080.002.001 FG.081.001.001 FG.081.002.001
## 1 2 1 2 1
## FG.082.001.001 FG.082.002.001 FG.083.001.001 FG.083.002.001 FG.083.003.001
## 1 1 1 1 1
## FG.084.001.001 FG.084.002.001 FG.084.003.001 FG.085.001.001 FG.085.001.002
## 1 1 1 1 1
## FG.085.001.003 FG.085.002.001 FG.086.001.001 FG.086.001.002 FG.086.002.001
## 1 1 1 1 1
## FG.087.001.001 FG.087.002.001 FG.087.002.002 FG.088.001.001 FG.088.002.001
## 2 1 1 1 1
## FG.089.001.001 FG.089.002.001 FG.090.001.001 FG.090.002.001 FG.091.001.001
## 2 1 1 1 2
## FG.092.001.001 FG.093.001.001 FG.093.001.002 FG.094.001.001 FG.094.002.001
## 2 1 1 2 1
## FG.095.001.001 FG.095.001.002 FG.096.001.001 FG.096.002.001 FG.097.001.001
## 2 1 3 1 1
## FG.097.002.001 FG.098.001.001 FG.098.002.001 FG.098.003.001 FG.099.001.001
## 1 1 1 1 2
## FG.099.002.001 FG.099.003.001 FG.100.001.001 FG.100.002.001 FG.101.001.001
## 1 1 2 1 2
## FG.101.001.002 FG.102.001.001 FG.102.002.001 FG.102.003.001 FG.103.001.001
## 1 1 1 1 2
## FG.104.001.001 FG.104.002.001 FG.105.001.001 FG.105.002.001 FG.106.001.001
## 1 1 2 1 2
## FG.107.001.001 FG.107.001.002 FG.107.002.001 FG.108.001.001 FG.108.002.001
## 1 1 1 1 1
## FG.109.001.001 FG.109.002.001 FG.110.001.001 FG.111.001.001 FG.112.001.001
## 1 1 2 2 1
## FG.112.002.001 FG.113.001.001 FG.114.001.001 FG.115.001.001 FG.116.001.001
## 1 1 1 1 1
## FG.117.001.001 FG.118.001.001
## 1 1In many cases, pre-defined feature groups (by the retention time
similarity and abundance correlation) were not further subdivided. Below
we evaluate some of the feature groups, starting with
FG.008.001 which was split into two different feature groups
based on EIC correlation. We first extract the EICs for all features
from this initial feature group. With n = 1 we specify to
extract the EIC only from the sample with the highest intensity.
fidx <- grep("FG.013.001.", featureGroups(xmse))
eics <- featureChromatograms(
xmse, features = rownames(featureDefinitions(xmse))[fidx],
filled = TRUE, n = 1)Next we plot the EICs using a different color for each of the
subgroups. With peakType = "none" we disable the
highlighting of the detected chromatographic peaks.
cols <- c("#ff000080", "#0000ff80")
names(cols) <- unique(featureGroups(xmse)[fidx])
plotChromatogramsOverlay(eics, col = cols[featureGroups(xmse)[fidx]],
lwd = 2, peakType = "none")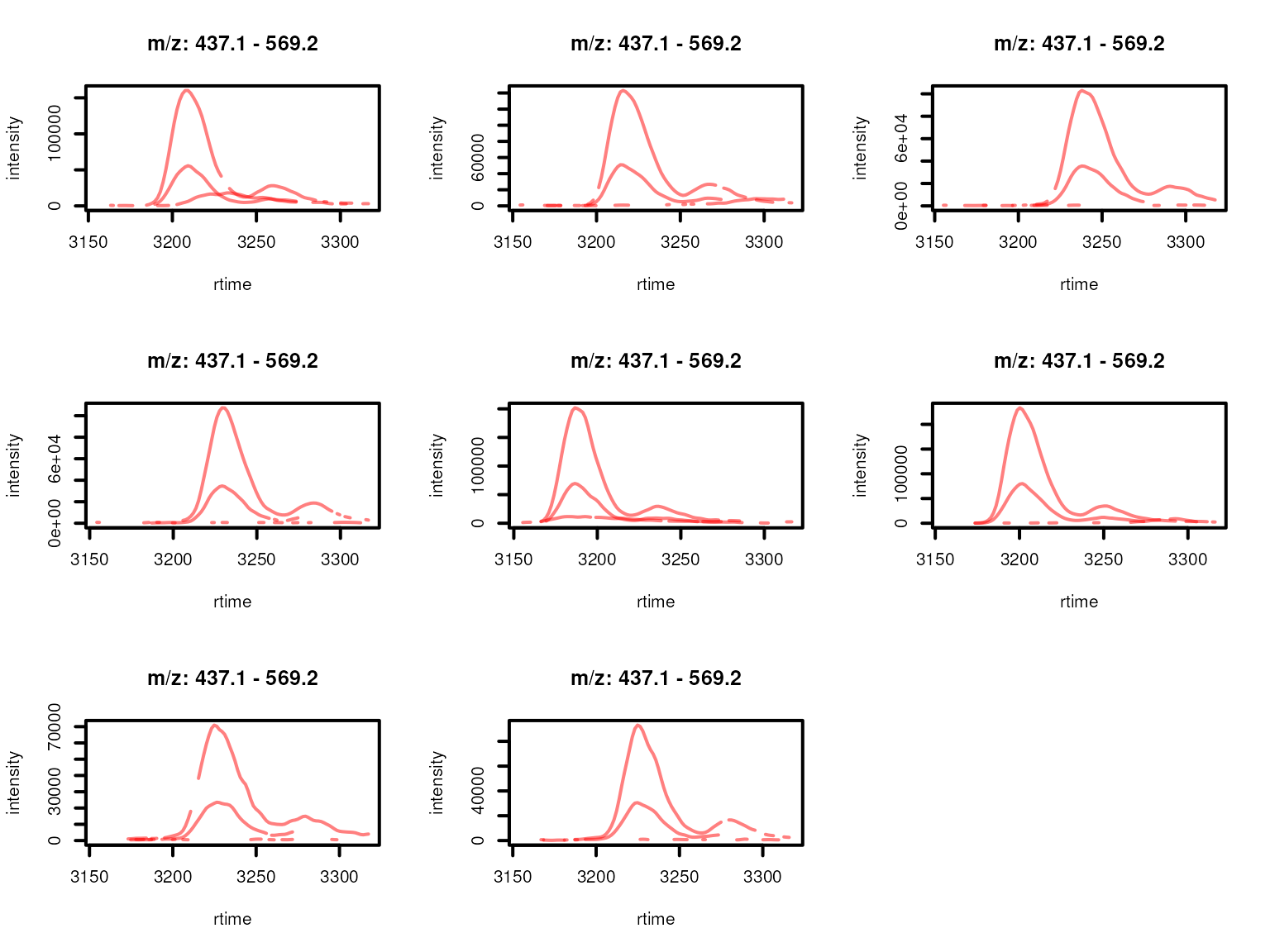
Feature EICs per sample for features from a feature group defined by rentention time and feature abudances across samples. Features with high correlation of their EICs are shown in the same color.
plotChromatogramsOverlay(normalize(eics),
col = cols[featureGroups(xmse)[fidx]],
lwd = 2, peakType = "none")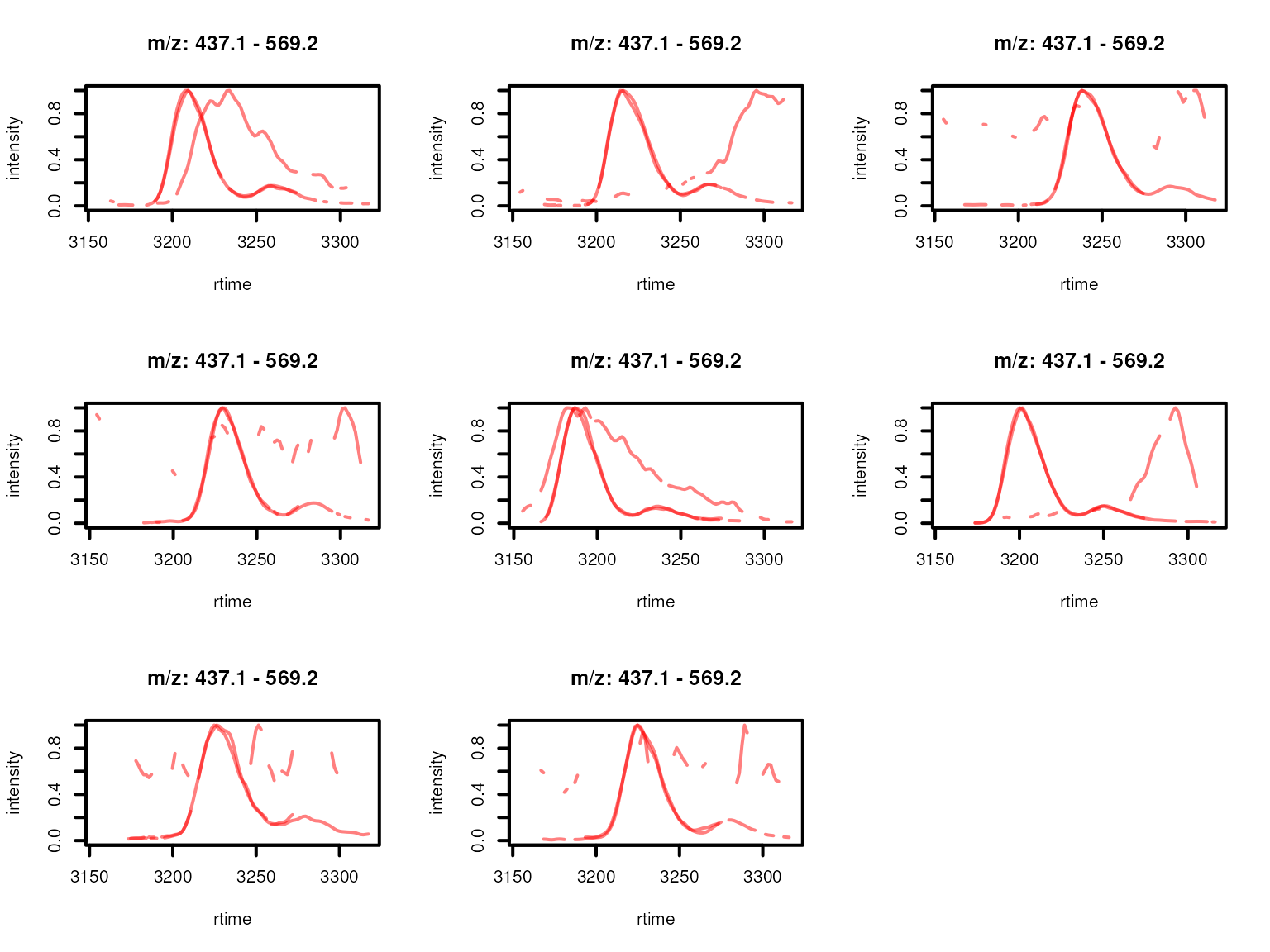
Feature EICs per sample normalized to an absolute intensity of 1 for features from a feature group defined by rentention time and feature abudances across samples. Features with high correlation of their EICs are shown in the same color.
One of the features (highlighted in red in the plots above) within the original feature group was separated from the other two because of a low similarity of their EICs. In fact, the feature’s EIC is shifted in some samples along the retention time dimension and can thus not represent the signal from the same compound.
We evaluate next the sub-grouping in another feature group.
fidx <- grep("FG.045.001.", featureGroups(xmse))
eics <- featureChromatograms(
xmse, features = rownames(featureDefinitions(xmse))[fidx],
filled = TRUE, n = 1)Next we plot the EICs using a different color for each of the subgroups.
cols <- c("#ff000080", "#0000ff80")
names(cols) <- unique(featureGroups(xmse)[fidx])
plotChromatogramsOverlay(eics, col = cols[featureGroups(xmse)[fidx]],
lwd = 2, peakType = "none")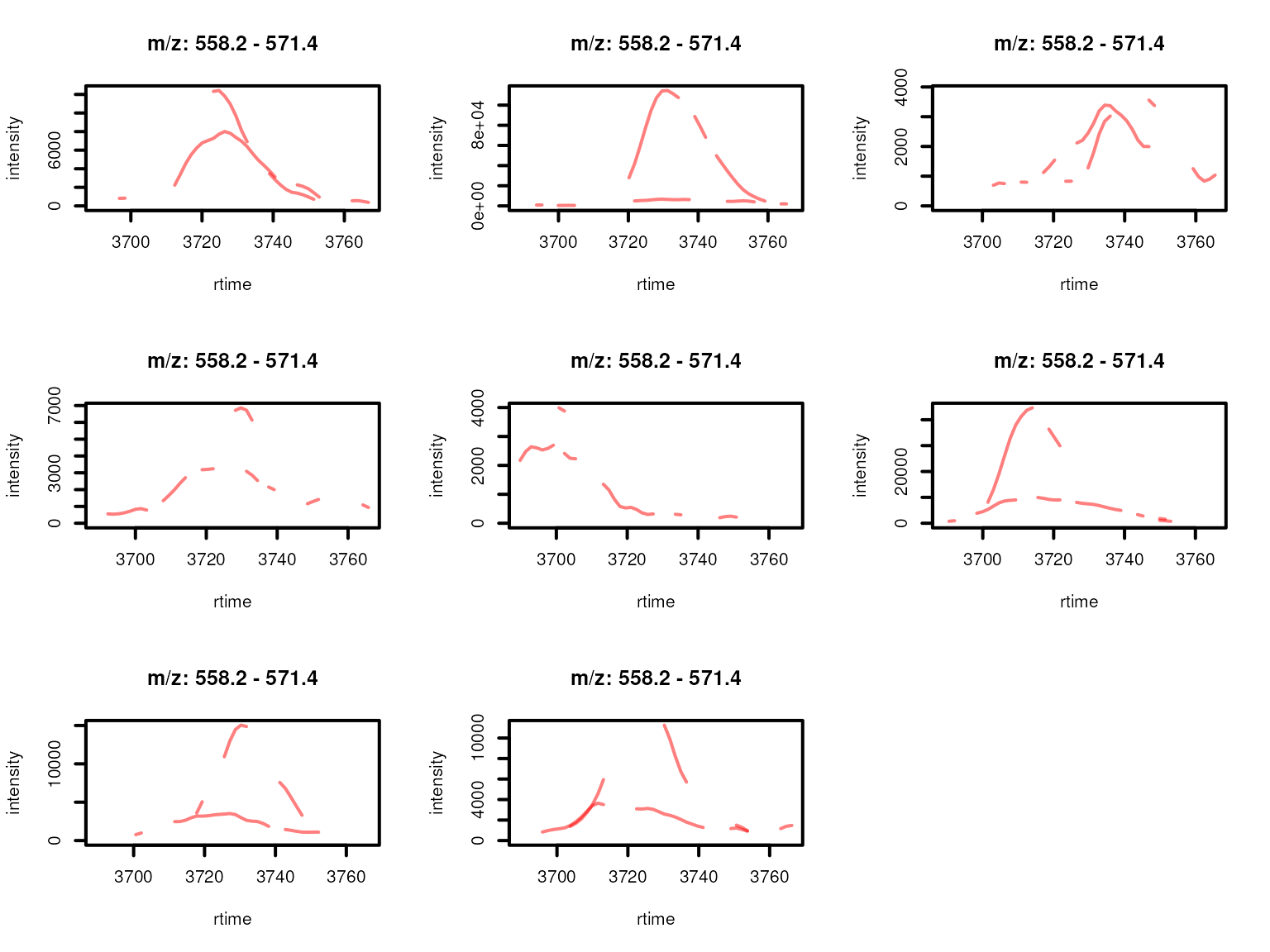
Feature EICs per sample for features from a feature group defined by rentention time and feature abudances across samples. Features with high correlation of their EICs are shown in the same color.
plotChromatogramsOverlay(normalize(eics),
col = cols[featureGroups(xmse)[fidx]],
lwd = 2, peakType = "none")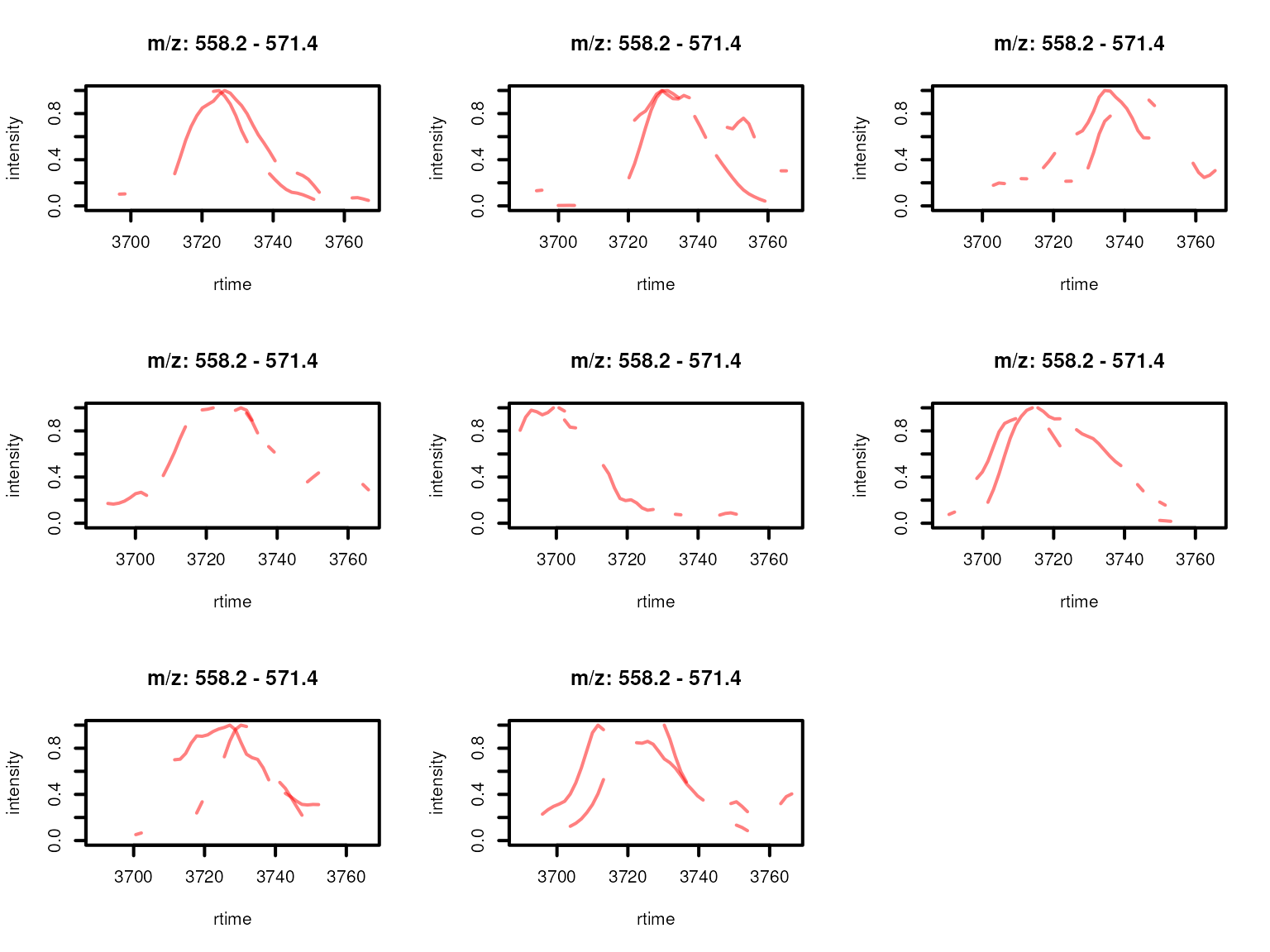
Feature EICs per sample normalized to an absolute intensity of 1 for features from a feature group defined by rentention time and feature abudances across samples. Features with high correlation of their EICs are shown in the same color.
Based on the EIC correlation, the initial feature group FG.045.001 was grouped into two separate sub-groups.
The grouping based on EIC correlation on the pre-defined feature groups from the previous sections grouped the 351 features into 267 feature groups.
Grouping of subsets of features
In the previous sections we were always considering all features from
the data set, but sometimes it could be desirable to just group a
pre-defined set of features, for example features found to be of
particular interest in a certain experiment (e.g. significant features).
This can be easily achieved by assigning the features of interest to a
initial feature group, using NA as group ID for all other
features.
To illustrate this we reset all feature groups by setting
them to NA and assign our features of interest (in this
example just 30 randomly selected features) to an initial feature group
"FG".
featureDefinitions(xmse)$feature_group <- NA_character_
set.seed(123)
fts_idx <- sample(1:nrow(featureDefinitions(xmse)), 30)
featureDefinitions(xmse)$feature_group[fts_idx] <- "FG"Any call to groupFeatures would now simply sub-group
this set of 30 features. Any feature which has an NA in the
"feature_group" column will be ignored.
xmse <- groupFeatures(xmse, SimilarRtimeParam(diffRt = 20))
xmse <- groupFeatures(xmse, AbundanceSimilarityParam(threshold = 0.7))
table(featureGroups(xmse))##
## FG.001.001 FG.001.002 FG.002.001 FG.002.002 FG.003.001 FG.003.002 FG.004.001
## 1 1 1 1 1 1 1
## FG.004.002 FG.005.001 FG.006.001 FG.006.002 FG.006.003 FG.007.001 FG.007.002
## 1 2 1 1 1 2 1
## FG.008.001 FG.009.001 FG.010.001 FG.011.001 FG.012.001 FG.013.001 FG.014.001
## 1 1 1 1 1 1 1
## FG.015.001 FG.016.001 FG.017.001 FG.018.001 FG.019.001 FG.020.001 FG.021.001
## 1 1 1 1 1 1 1Session information
## R Under development (unstable) (2026-02-14 r89420)
## Platform: x86_64-pc-linux-gnu
## Running under: Ubuntu 24.04.3 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## time zone: UTC
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] pheatmap_1.0.13 faahKO_1.51.0 MSnbase_2.37.0
## [4] ProtGenerics_1.43.0 S4Vectors_0.49.0 mzR_2.45.0
## [7] Rcpp_1.1.1 Biobase_2.71.0 BiocGenerics_0.57.0
## [10] generics_0.1.4 MsFeatures_1.19.0 xcms_4.9.2
## [13] BiocParallel_1.45.0 BiocStyle_2.39.0
##
## loaded via a namespace (and not attached):
## [1] DBI_1.2.3 rlang_1.1.7
## [3] magrittr_2.0.4 clue_0.3-67
## [5] MassSpecWavelet_1.77.0 otel_0.2.0
## [7] matrixStats_1.5.0 compiler_4.6.0
## [9] systemfonts_1.3.1 vctrs_0.7.1
## [11] reshape2_1.4.5 stringr_1.6.0
## [13] crayon_1.5.3 pkgconfig_2.0.3
## [15] MetaboCoreUtils_1.19.2 fastmap_1.2.0
## [17] XVector_0.51.0 rmarkdown_2.30
## [19] preprocessCore_1.73.0 ragg_1.5.0
## [21] purrr_1.2.1 xfun_0.56
## [23] MultiAssayExperiment_1.37.2 cachem_1.1.0
## [25] jsonlite_2.0.0 progress_1.2.3
## [27] DelayedArray_0.37.0 prettyunits_1.2.0
## [29] parallel_4.6.0 cluster_2.1.8.2
## [31] R6_2.6.1 bslib_0.10.0
## [33] stringi_1.8.7 RColorBrewer_1.1-3
## [35] limma_3.67.0 GenomicRanges_1.63.1
## [37] jquerylib_0.1.4 iterators_1.0.14
## [39] Seqinfo_1.1.0 bookdown_0.46
## [41] SummarizedExperiment_1.41.1 knitr_1.51
## [43] IRanges_2.45.0 Matrix_1.7-4
## [45] igraph_2.2.2 tidyselect_1.2.1
## [47] abind_1.4-8 yaml_2.3.12
## [49] doParallel_1.0.17 codetools_0.2-20
## [51] affy_1.89.0 lattice_0.22-9
## [53] tibble_3.3.1 plyr_1.8.9
## [55] S7_0.2.1 evaluate_1.0.5
## [57] desc_1.4.3 Spectra_1.21.1
## [59] pillar_1.11.1 affyio_1.81.0
## [61] BiocManager_1.30.27 MatrixGenerics_1.23.0
## [63] foreach_1.5.2 MALDIquant_1.22.3
## [65] ncdf4_1.24 hms_1.1.4
## [67] ggplot2_4.0.2 scales_1.4.0
## [69] MsExperiment_1.13.1 glue_1.8.0
## [71] lazyeval_0.2.2 tools_4.6.0
## [73] mzID_1.49.0 data.table_1.18.2.1
## [75] QFeatures_1.21.0 vsn_3.79.5
## [77] fs_1.6.6 XML_3.99-0.22
## [79] grid_4.6.0 impute_1.85.0
## [81] tidyr_1.3.2 MsCoreUtils_1.23.2
## [83] PSMatch_1.15.1 cli_3.6.5
## [85] textshaping_1.0.4 S4Arrays_1.11.1
## [87] dplyr_1.2.0 AnnotationFilter_1.35.0
## [89] pcaMethods_2.3.0 gtable_0.3.6
## [91] sass_0.4.10 digest_0.6.39
## [93] SparseArray_1.11.10 htmlwidgets_1.6.4
## [95] farver_2.1.2 htmltools_0.5.9
## [97] pkgdown_2.2.0.9000 lifecycle_1.0.5
## [99] statmod_1.5.1 MASS_7.3-65