centWave-based peak detection in purely chromatographic data
Source:R/methods-Chromatogram.R, R/methods-MChromatograms.R
findChromPeaks-Chromatogram-CentWaveParam.RdfindChromPeaks on a MSnbase::Chromatogram or MSnbase::MChromatograms
object with a CentWaveParam parameter object performs centWave-based
peak detection on purely chromatographic data. See centWave for details
on the method and CentWaveParam for details on the parameter class.
Note that not all settings from the CentWaveParam will be used.
See peaksWithCentWave() for the arguments used for peak detection
on purely chromatographic data.
After chromatographic peak detection, identified peaks can also be refined
with the refineChromPeaks() method, which can help to reduce peak
detection artifacts.
Usage
# S4 method for class 'Chromatogram,CentWaveParam'
findChromPeaks(object, param, ...)
# S4 method for class 'MChromatograms,CentWaveParam'
findChromPeaks(object, param, BPPARAM = bpparam(), ...)
# S4 method for class 'MChromatograms,MatchedFilterParam'
findChromPeaks(object, param, BPPARAM = BPPARAM, ...)Arguments
- object
a MSnbase::Chromatogram or MSnbase::MChromatograms object.
- param
a CentWaveParam object specifying the settings for the peak detection. See
peaksWithCentWave()for the description of arguments used for peak detection.- ...
currently ignored.
- BPPARAM
a parameter class specifying if and how parallel processing should be performed (only for
XChromatogramsobjects). It defaults tobpparam(). SeeBiocParallel::bpparam()for more information.
Value
If called on a Chromatogram object, the method returns an XChromatogram
object with the identified peaks. Columns "mz", "mzmin" and "mzmax" in
the chromPeaks() peak matrix provide the mean m/z and the maximum and
minimum m/z value of the Chromatogram object. See peaksWithCentWave()
for details on the remaining columns.
See also
peaksWithCentWave() for the downstream function and centWave
for details on the method.
Examples
library(MSnbase)
## Loading a test data set with identified chromatographic peaks
faahko_sub <- loadXcmsData("faahko_sub2")
faahko_sub <- filterRt(faahko_sub, c(2500, 3700))
#> Filter spectra
##
od <- as(filterFile(faahko_sub, 1L), "MsExperiment")
## Extract chromatographic data for a small m/z range
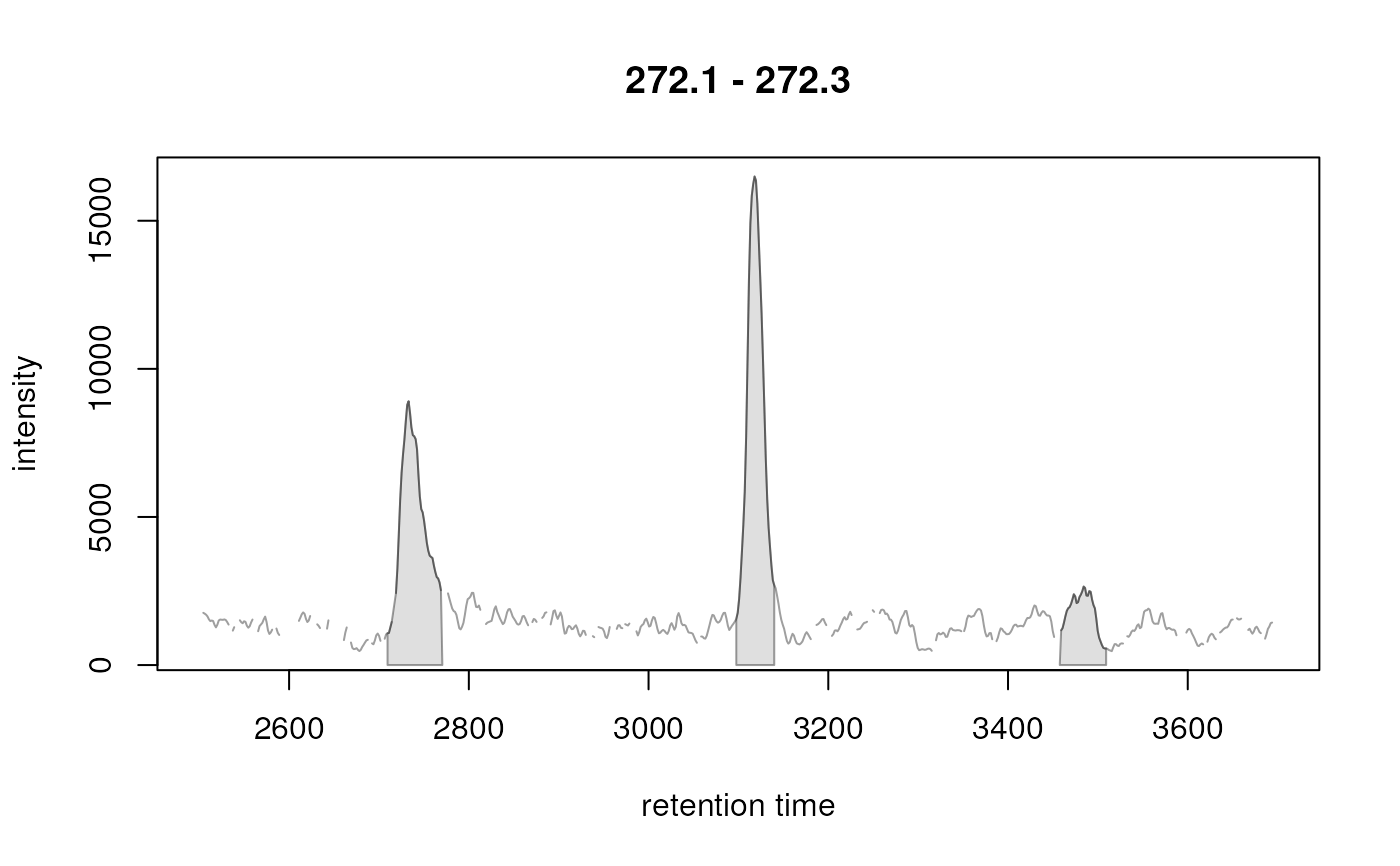
chr <- chromatogram(od, mz = c(272.1, 272.3))[1, 1]
## Identify peaks with default settings
xchr <- findChromPeaks(chr, CentWaveParam())
xchr
#> Object of class: XChromatogram
#> Intensity values aggregated using: sum
#> length of object: 766
#> from file: 1
#> mz range: [272.1, 272.3]
#> rt range: [2501.378, 3698.567]
#> MS level: 1
#> Identified chromatographic peaks (3):
#> rt rtmin rtmax into maxo sn (4 more column(s))
#> 2734.556 2709.517 2770.55 286653.221615386 8907 15
#> 3117.97 3097.625 3139.879 372049.599925925 16496 33
#> 3487.299 3457.565 3509.208 93049.7314242425 2651 15
## Plot data and identified peaks.
plot(xchr)
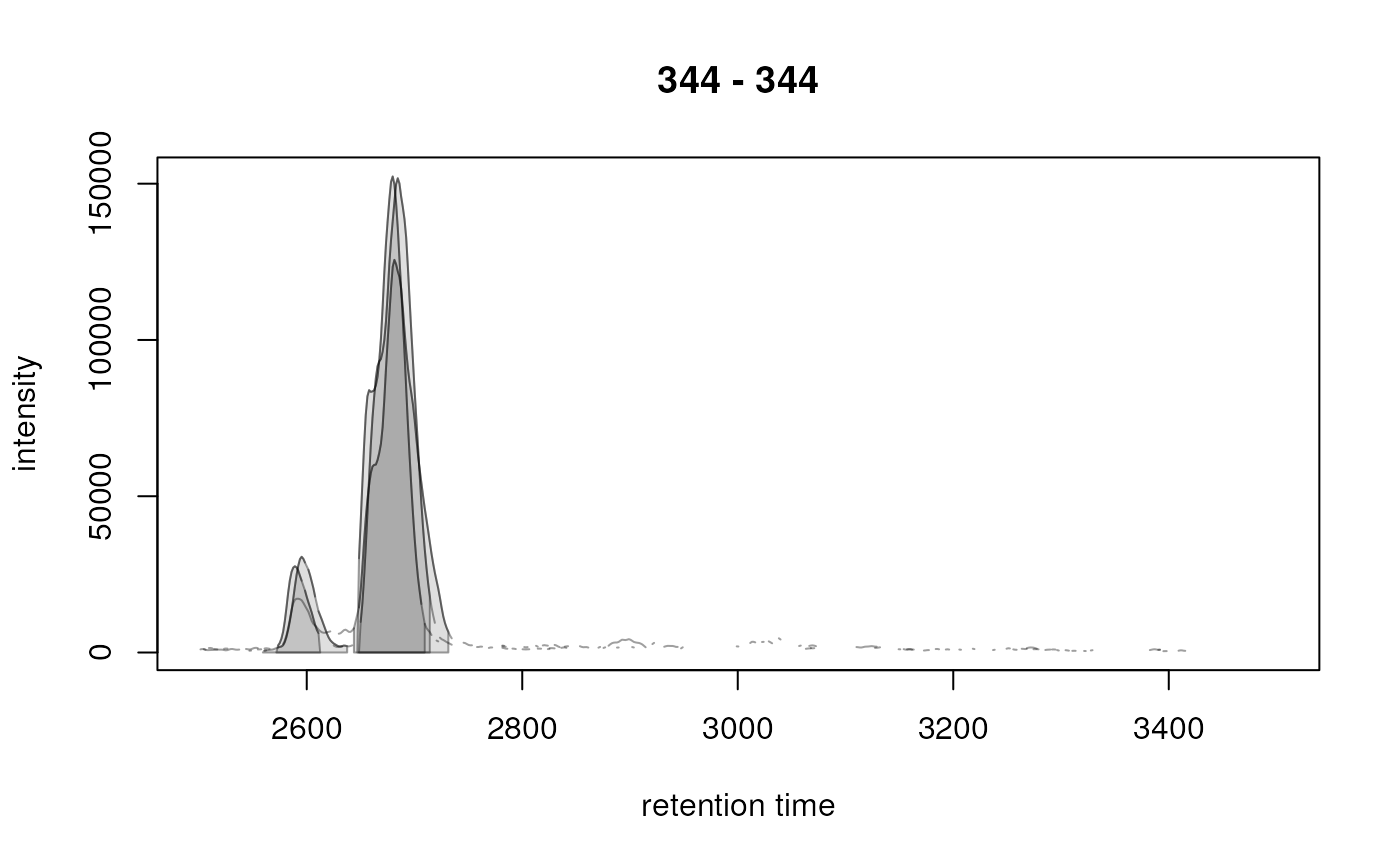
 library(MsExperiment)
library(xcms)
## Perform peak detection on an MChromatograms object
fls <- c(system.file("cdf/KO/ko15.CDF", package = "faahKO"),
system.file("cdf/KO/ko16.CDF", package = "faahKO"),
system.file("cdf/KO/ko18.CDF", package = "faahKO"))
od3 <- readMsExperiment(fls)
## Disable parallel processing for this example
register(SerialParam())
## Extract chromatograms for a m/z - retention time slice
chrs <- chromatogram(od3, mz = 344, rt = c(2500, 3500))
## Perform peak detection using CentWave
xchrs <- findChromPeaks(chrs, param = CentWaveParam())
xchrs
#> XChromatograms with 1 row and 3 columns
#> ko15.CDF ko16.CDF ko18.CDF
#> <XChromatogram> <XChromatogram> <XChromatogram>
#> [1,] peaks: 2 peaks: 2 peaks: 1
#> phenoData with 2 variables
#> featureData with 4 variables
#> - - - xcms preprocessing - - -
#> Chromatographic peak detection:
#> method: centWave
## Extract the identified chromatographic peaks
chromPeaks(xchrs)
#> mz mzmin mzmax rt rtmin rtmax into intb maxo sn
#> mzmin 344 344 344 2589.015 2571.801 2612.490 592297.3 554533.9 27600 14
#> mzmin 344 344 344 2679.783 2646.919 2709.517 5210015.9 5071346.6 152320 30
#> mzmin 344 344 344 2593.710 2559.281 2637.529 722415.3 640838.6 30640 14
#> mzmin 344 344 344 2686.042 2648.483 2714.211 5700652.0 5587464.2 151744 45
#> mzmin 344 344 344 2682.914 2643.790 2731.427 5255689.5 4983388.9 125632 28
#> row column
#> mzmin 1 1
#> mzmin 1 1
#> mzmin 1 2
#> mzmin 1 2
#> mzmin 1 3
## plot the result
plot(xchrs)
library(MsExperiment)
library(xcms)
## Perform peak detection on an MChromatograms object
fls <- c(system.file("cdf/KO/ko15.CDF", package = "faahKO"),
system.file("cdf/KO/ko16.CDF", package = "faahKO"),
system.file("cdf/KO/ko18.CDF", package = "faahKO"))
od3 <- readMsExperiment(fls)
## Disable parallel processing for this example
register(SerialParam())
## Extract chromatograms for a m/z - retention time slice
chrs <- chromatogram(od3, mz = 344, rt = c(2500, 3500))
## Perform peak detection using CentWave
xchrs <- findChromPeaks(chrs, param = CentWaveParam())
xchrs
#> XChromatograms with 1 row and 3 columns
#> ko15.CDF ko16.CDF ko18.CDF
#> <XChromatogram> <XChromatogram> <XChromatogram>
#> [1,] peaks: 2 peaks: 2 peaks: 1
#> phenoData with 2 variables
#> featureData with 4 variables
#> - - - xcms preprocessing - - -
#> Chromatographic peak detection:
#> method: centWave
## Extract the identified chromatographic peaks
chromPeaks(xchrs)
#> mz mzmin mzmax rt rtmin rtmax into intb maxo sn
#> mzmin 344 344 344 2589.015 2571.801 2612.490 592297.3 554533.9 27600 14
#> mzmin 344 344 344 2679.783 2646.919 2709.517 5210015.9 5071346.6 152320 30
#> mzmin 344 344 344 2593.710 2559.281 2637.529 722415.3 640838.6 30640 14
#> mzmin 344 344 344 2686.042 2648.483 2714.211 5700652.0 5587464.2 151744 45
#> mzmin 344 344 344 2682.914 2643.790 2731.427 5255689.5 4983388.9 125632 28
#> row column
#> mzmin 1 1
#> mzmin 1 1
#> mzmin 1 2
#> mzmin 1 2
#> mzmin 1 3
## plot the result
plot(xchrs)